ROEBA
Row-by-row, Odd/even by amplifier correction
This is useful for JWST instruments where one want to improve upon the reference pixel step
[1]:
from tshirt.tests import test_phot_algorithms
import numpy as np
from tshirt.pipeline.instrument_specific import rowamp_sub
%matplotlib inline
Test Simualated Images
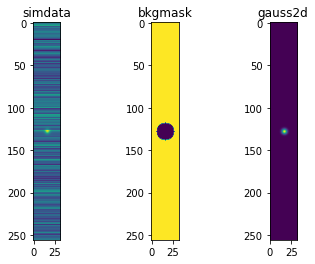
Simulate some “row-like” noise in an image that looks like 1/f noise. The simulated data has a noiseless star on top The following plots are * “simdata” - the simulated data * “bkgmask” - the mask of background pixels * “gauss2d” - the Gaussian star alone
[2]:
rab = test_phot_algorithms.rowAmpBacksub()
simDict = rab.sim_data()
rab.show_images()
/Users/everettschlawin/es_programs/tshirt/tshirt/tests/test_phot_algorithms.py:129: UserWarning: Matplotlib is currently using module://matplotlib_inline.backend_inline, which is a non-GUI backend, so cannot show the figure.
fig.show()
Now test the simulation with ROEBA applied
The plots are now * “simdata” - the simulated data * “bkgmask” - the mask of background pixels * “gauss2d” - the Gaussian star alone * “model” - the ROEBA model from background pixels * “resid” - the residual of the ROEBA-applied image and the original Gaussian 2D
[3]:
outimg, modelimg = rowamp_sub.do_backsub(simDict['simdata'],
amplifiers=1,
backgMask=simDict['bkgmask'])
simDict['model'] = modelimg
simDict['outimg'] = outimg
simDict['resid'] = simDict['gauss2d'] - simDict['outimg']
rab.show_images(simDict)
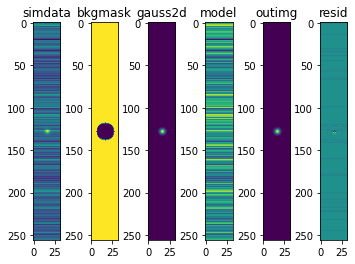
The residuals don’t look perfect. But further examination shows they are within machine errors
[4]:
np.max(np.abs((simDict['resid'])))
[4]:
4.440892098500626e-16
How to use it with the JWST pipeline?
You can run ROEBA after the superbias step like so. This assumes you have already run the dq init and saturation flagging steps and called the result saturation
from jwst.superbias import SuperBiasStep
superbias_step = SuperBiasStep()
# superbias_step.output_dir = output_dir
# superbias_step.save_results = True
# Call using the the output from the previously-run saturation step
superbias = superbias_step.run(saturation)
mod_refpix = deepcopy(superbias)
ngroups = superbias.meta.exposure.ngroups
nints = superbias.data.shape[0] ## could be split into ints per segment
for oneInt in tqdm.tqdm(np.arange(nints)):
for oneGroup in np.arange(ngroups):
rowSub, modelImg = rowamp_sub.do_backsub(superbias.data[oneInt,oneGroup,:,:],
backgMask=simDict['bkgmask'],amplifiers=1)
mod_refpix.data[oneInt,oneGroup,:,:] = rowSub
[ ]: